THE RESULTS
The PC-IM algorithm has been evaluated on expression data of the plant Arabidopsis Thaliana, using its flower organ specification gene regulatory network (FOS-GRN).
There have been three kind of evaluation:
1. PRELIMINARY EVALUATION
Expression data in silico (generated from mathematical equations) and in vivo (real data available in public databases) have been studied in order to find the most reliable ones. The in silico method showed greater precision and sensitivity, but its results were influenced by the algorithm used; whereas the in vivo method overcame this problem, and for this reason it has been chosen.
Also, the PC and ARACNE algorithm have been compared, to find the most effective one in the GRN expansion. The winner was the PC, since it has better performance when applied to real gene expression data and a better PPV.
2. PC-IM EVALUATION
Four sub-experiments has been performed to analyze the PC-IM algorithm:
a. Size of blocks: the algorithm has been run with blocks of different sizes. The optimal values were found by using 1000 genes
b. Number of iterations: nine different iteration values have been analyzed; the best performance have been obtained with 100 iterations
c. Robustness: the PC-IM has been run both with a FOS-GRN and with a non-real GRN as inputs. In the first case it has reached better PPV and sensitivity, showing that it is robust
d. Comparison with GENIES: the algorithm has been compared with GENIES, a competitor method recently developed for the LGN expansion. This one showed better expansion performance, but it did not find any extra gene. The result of the PC-IM was a larger number of genes in the final expansion list. Therefore, the PC-IM can be considered more efficient in the LGN expansion task
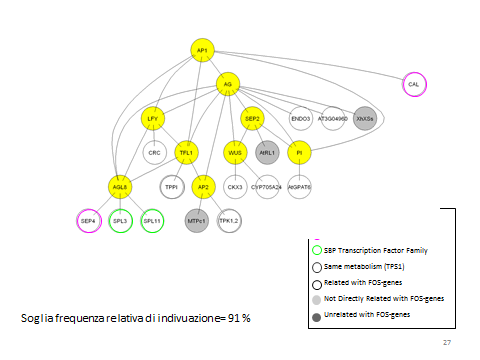
3. BIOLOGICAL VALIDATION
The final results of the PC-IM has been validated through a bibliographic search. This process found certain correlations or non-correlations for almost the 50% of genes; whereas for the remaining ones there were not useful references describing their functions. In conclusion, even though it is not possible to validate all the genes because some of them are not addressed in studies, those at the top of the expansion list are strongly related to the LGN.
Also, results have been evaluated compared to the ones obtained by using random genes. The output of the PC-IM was significant, contrary to that of the random ones (the LR+ value was very low). This means that the genes found by the PC-IM have good probabilities to be related with those of the LGN, and that have low probability to be randomly obtained.